Using AI to comprehend unstructured clinical documents
One of the most successful systems which I was responsible for architecting & developing has been ALMA, a clinic letters system used by a large NHS trust. It’s been in production for nine years and has produced over 3.5 million letters. On the whole its well designed and serves its use case daily.
However, there is one feature that has been misused by the hundreds of users over the years. The medical secretaries using the system requested a feature to add comments and notes to the patients record. This feature was intended for the medical secretaries to leave quick notes to each other about administrative tasks which still needed completing.
Over time, this commenting feature started being used by clinical staff to leave notes about suggested changes in medication. We now have the situation where we dont’t know for definite which comments are purely administrative and which ones contain clinical information and therefore should be included in the shared care record.
AWS Medical Comprehend to the rescue?
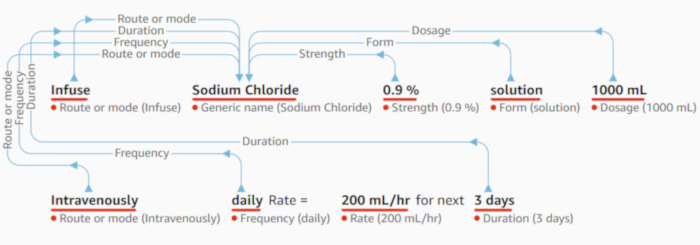
The medical comprehend API is a recent offering from AWS which accepts unstructured medical text and returns a JSON object outlining all the medical, procedural and diagnosis terms found within the text. For medications it also returns the dose, route and frequency if found within the text. For each medical term found a confidence score is returned to help you decided if you should trust the value. An offset position is also returned indicating the starting point for the medical term found.
Using this API it would be possible to process all the patient comments and if enough medical terms are detected then we can flag the comment as having Clinical content.
Many hospitals still produce large quantities of unstructured clinical data as PDF formatted reports. If budgets permit, its possible to post process these PDFs after publication to add a medical term search index to the document.
Sounds expensive!
The current cost is $0.01 per work unit. A work unit is 100 characters of text. If a typical comment contains 300 characters and you need to process 10,000 comments the cost would be $300. There is a minimum charge for one work unit, so if a comment only has 60 characters of text you’ll still be charged one work unit. You could combine lots of smaller comments together into a larger work unit as long as you keep track of the start/end offsets for each comment. As the API returns the offsets of the detected terms you could then re-associate them with the originating comment.
Example using Go
The first step is to install the AWS software development kit (SDK) for Go. This is done by using the following Go get command issued at the terminal or command prompt.
go get github.com/aws/aws-sdk-go/...
Once the AWS SDK has been installed, you’ll then need to import the relevant sections into your program to be able to interact with the medical comprehend API.
package main
import (
"bytes"
"fmt"
"log"
"net/http"
"os"
"github.com/aws/aws-sdk-go/aws"
"github.com/aws/aws-sdk-go/aws/credentials"
"github.com/aws/aws-sdk-go/aws/session"
"github.com/aws/aws-sdk-go/service/comprehendmedical"
)
You’ll need to create an AWS session select a region which supports the medical comprehend API (not all do). In the example below the newly created session is assigned to the s variable. The session object is created by passing in the region identifier and your AWS credentials for id and secret key. Your id and secret key can be obtained from your AWS account.
func main() {
// create an AWS session which can be
// reused if we're uploading many files
s, err := session.NewSession(&aws.Config{
Region: aws.String("us-east-1"),
Credentials: credentials.NewStaticCredentials(
"XXX",
"YYY",
""),
})
if err != nil {
log.Fatal(err)
}
client := comprehendmedical.New(s)
input := comprehendmedical.DetectEntitiesInput{}
input.SetText(`
Pt is 40yo mother, highschool teacher
HPI : Sleeping trouble on present dosage of Clonidine.
Severe Rash on face and leg, slightly itchy
Meds : Vyvanse 50 mgs po at breakfast daily,
Clonidine 0.2 mgs -- 1 and 1 / 2 tabs po qhs
HEENT : Boggy inferior turbinates, No oropharyngeal lesion
Lungs : clear
Heart : Regular rhythm
Skin : Mild erythematous eruption to hairline
`)
result, err := client.DetectEntities(&input)
if err != nil {
log.Fatal(err)
}
}
Start by defining a medical comprehend client using the New function passing in your AWS session variable s. Then define an input object and use the SetText function to define the medical text you want to process. The processing of the medical text is done when the DetectEntities function is called and the results are stored in the results variable.
To get the JSON string returned you can use the GoString function as shown below. This is useful if you want to store the JSON in a Postgres JSONB database field to perform search queries across many patients.
fmt.Println(result.GoString())
You can loop over the detected terms and only print out the medications found by using the following pattern:
for _, entity := range result.Entities {
if *entity.Category == "MEDICATION" {
fmt.Println(*entity.Text)
fmt.Println(*entity.Type)
fmt.Println(*entity.Score)
fmt.Println("-----------")
}
}
Which will output the following results:
Clonidine
GENERIC_NAME
0.9948381781578064
-----------
Vyvanse
BRAND_NAME
0.9995348453521729
-----------
Clonidine
GENERIC_NAME
0.997945249080658
-----------
Categories
Other useful category names include DX_NAME, DIAGNOSIS, SYMPTOM, SYSTEM_ORGAN_SITE, DOSAGE & DIRECTION.
For a complete list of entity categories consult the documentation.
Languages supported
Amazon Comprehend can examine and analyse documents in these languages:
- English
- Spanish
- French
- German
- Italian
- Portuguese
